UGENE Tips: Pairwise Alignment, Remove Gaps and more
Pairwise Alignment
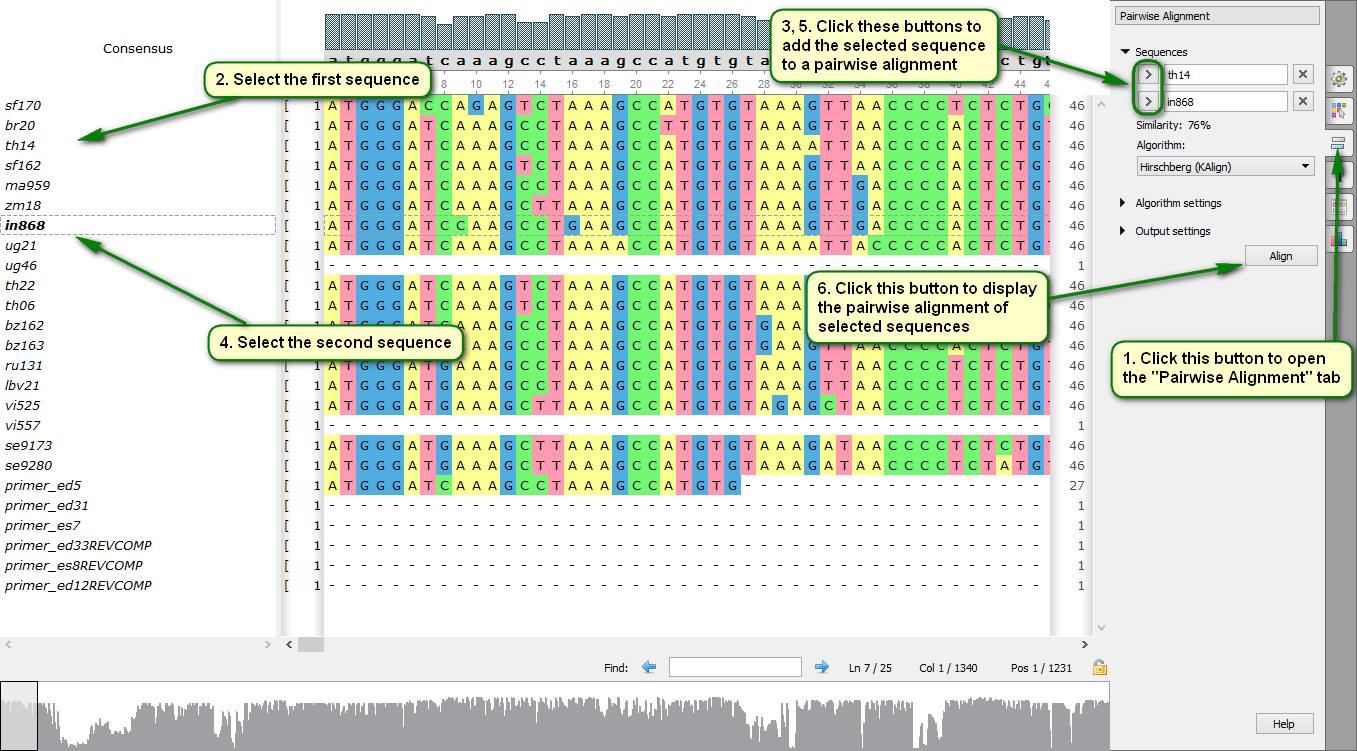
Did you know how to build quickly a pairwise alignment for sequences included to a multiple alignment?
To do this you need to open the "Pairwise Alignment" tab in the Alignment Editor Options Panel and specify two sequences there. The following algorithms are available for producing a pairwise alignment – Smith-Waterman algorithm and KAlign algorithm. Pairwise alignment results are saved to a separate file by default, but they also can be saved into the original file.
On attached screenshots you can see an example of how to build a pairwise alignment displayed in a separate window.
See more about the work with pairwise alignments in our documentation.
Distance
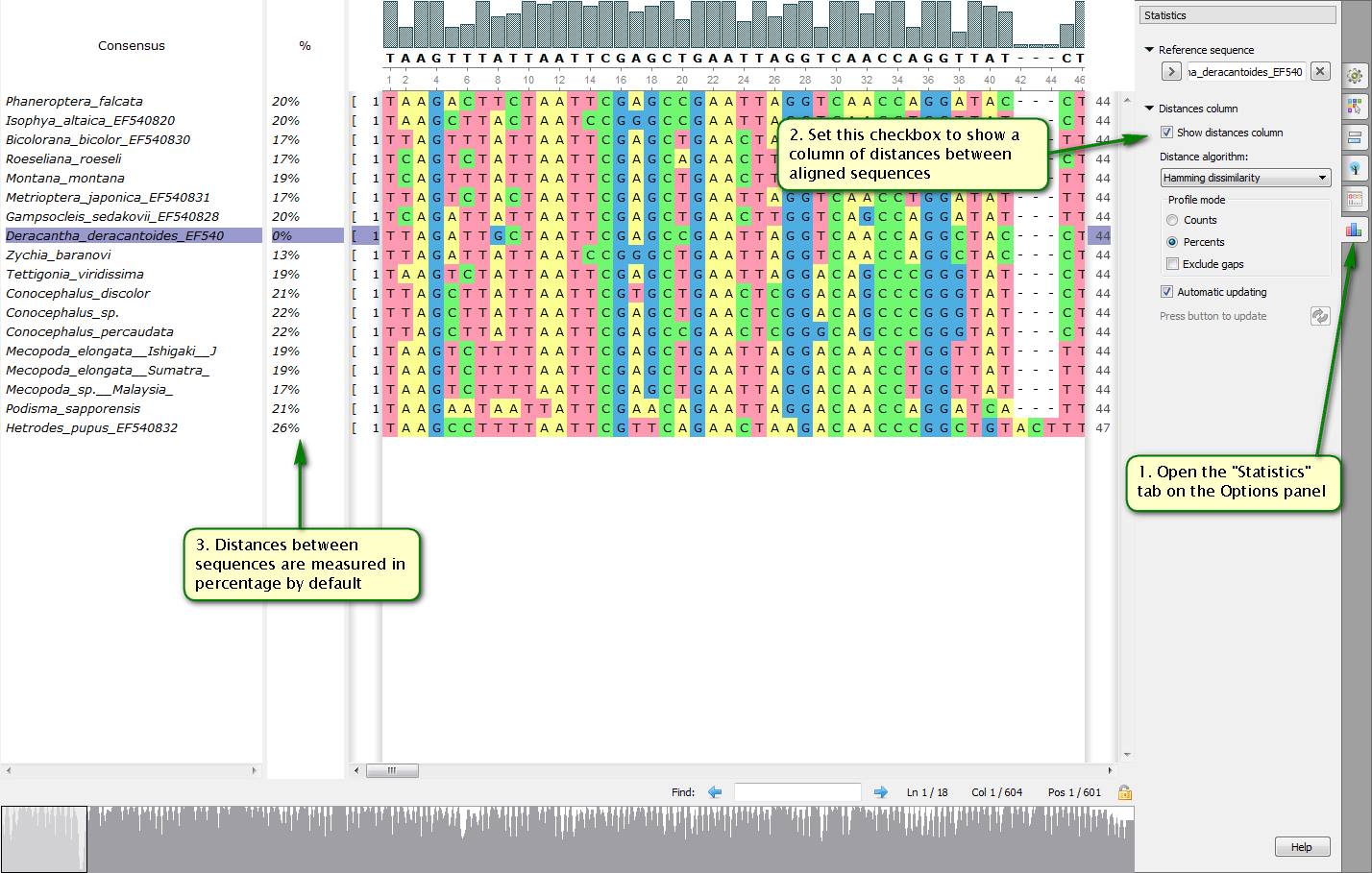
Did you know how to get quickly distance statistics about sequences included to a multiple alignment?
To do this you need to start with setting a reference sequence which others are going to be compared with. Then you should open the "Statistics" tab on the Options panel and set the "Show distances column" checkbox. After that your aligned sequences list is supplemented with a distance values column.
On attached screenshots you can see an example of how to show distances for a chosen reference sequence.
See more about displaying sequence statistics in the Multiple alignment editor in our documentation.
Remove Gaps
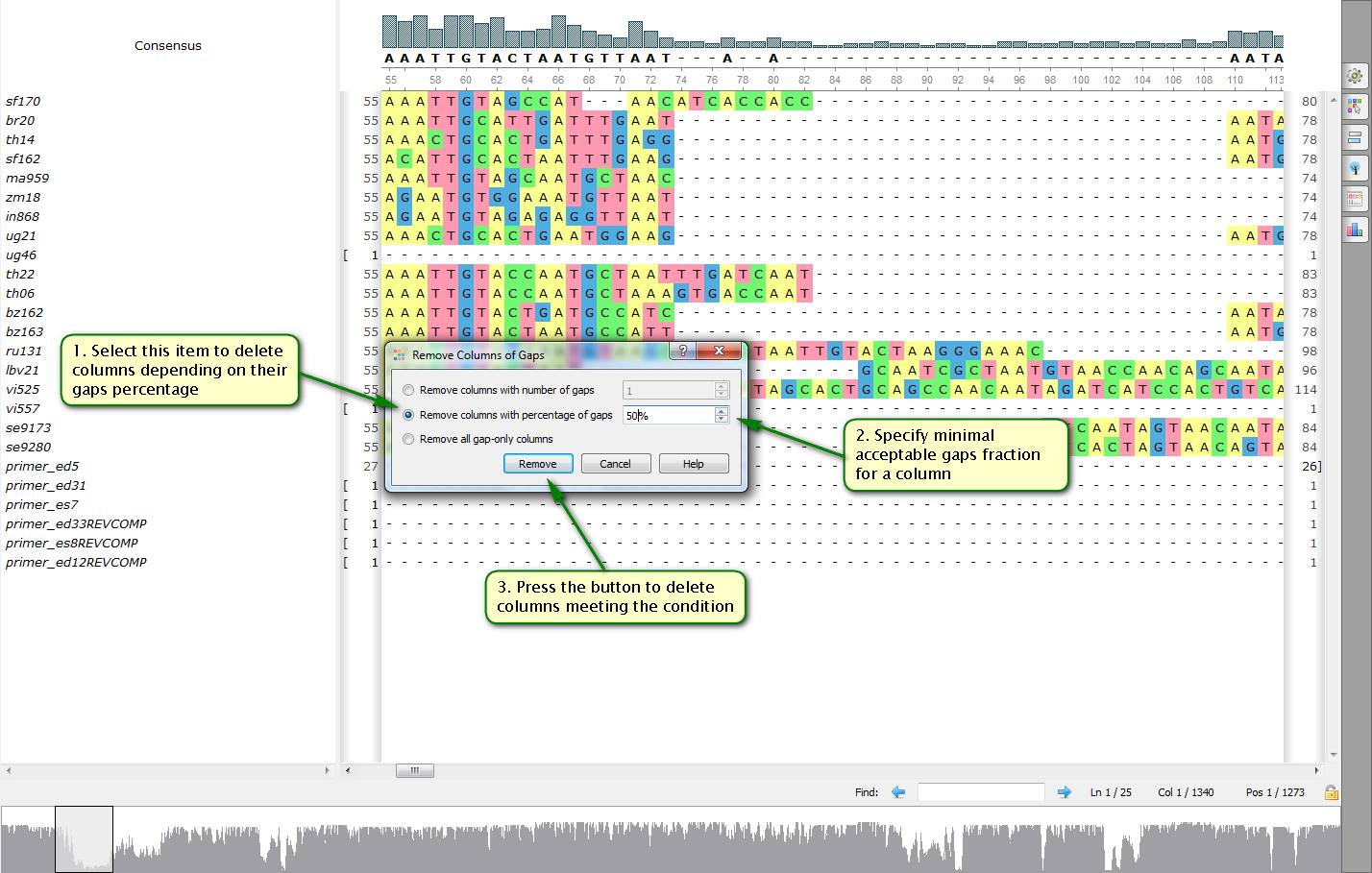
Did you know how to delete swiftly columns with gaps from a multiple alignment?
To do this open a needed file with a multiple alignment by UGENE. Then select "Edit -> Remove columns of gaps…" in the right-click menu. After that a dialog appears allowing you to specify a number of gaps in columns to be deleted. This number can be either an absolute one or a percentage of a bases count in a column.
Also there is a possibility of removing all the gaps within an alignment. In order to do this select "Edit -> Remove all gaps" in the multiple alignment editor right-click menu.
On attached screenshots you can see an example of how to delete columns more than a half of which consist of gaps from an alignment.
See more about deleting gaps from an alignment in our documentation.
Make a Phylogenetic Tree
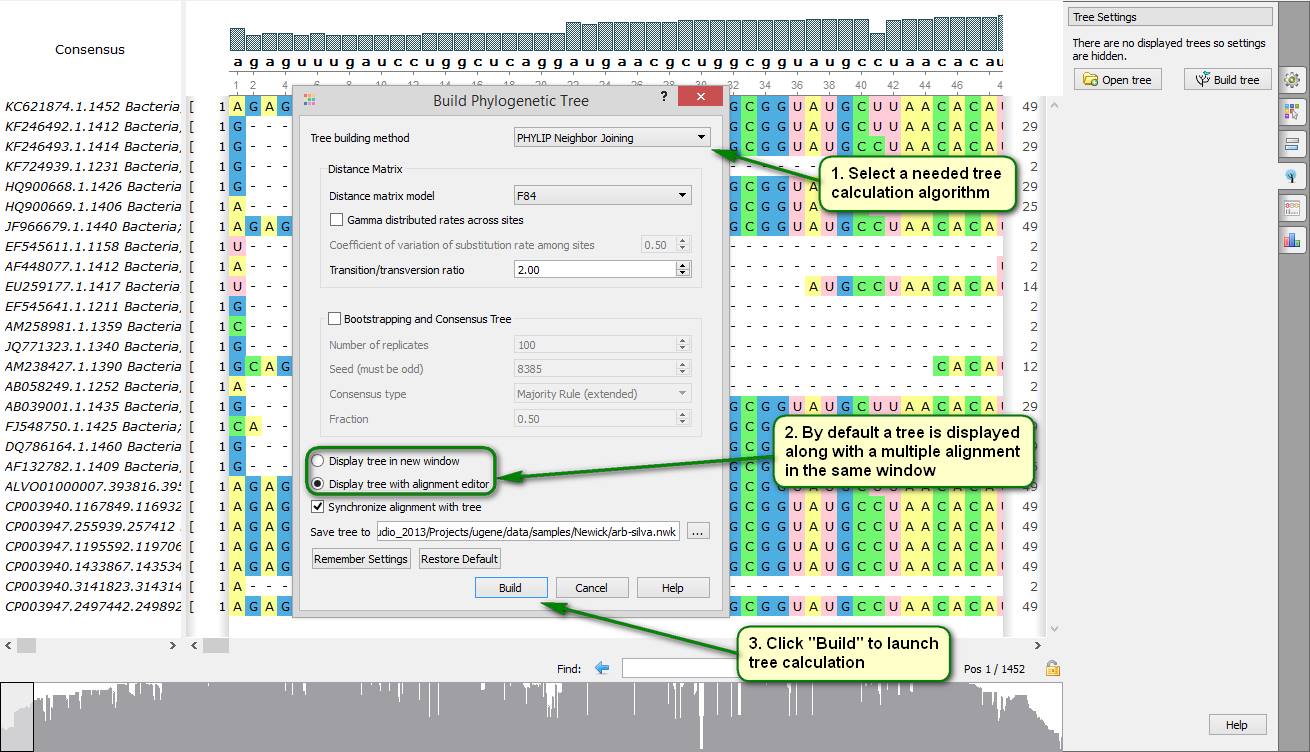
Did you know how to make a phylogenetic tree by a given multiple alignment?
To do this open a needed file with a multiple alignment by UGENE. Then expand the "Tree Settings" tab on the Options panel and click "Build tree". Then a dialog should appear in which you can specify a tree calculation algorithm and a path to a file which the tree is saved in. By default the tree is displayed in the same window as the multiple alignment. Alternatively, it can be shown in a separate window.
On attached screenshots you can see an example of how to make a phylogenetic tree for aligned 16S rRNA sequences of several strains of cyanobacteria.
See more about available tree calculation algorithms in our documentation.

