Performers assembly of input short reads.
Element type: spades-id
Parameters
| Parameter | Description | Default value | Parameter in Workflow File | Type |
|---|---|---|---|---|
| Input data | Select the type of input for SPAdes. URL(s) to the input files of the selected type(s) should be provided to the corresponding port(s) of the workflow element. At least one library of the following types is required:
It is strongly suggested to provide multiple paired-end and mate-pair libraries according to their insert size (from smallest to longest). Additionally, one may input Oxford Nanopore reads, Sanger reads, contigs generated by other assembler(s), etc. Note that Illumina and IonTorrent libraries should not be assembled together. All other types of input data are compatible. It is also possible to set up reads orientation (forward-reverse (fr), reverse-forward (rf), forward-forward (ff)) and specify whether paired reads are separate or interlaced. Illumina, IonTorrent or PacBio CCS reads should be provided in FASTQ format. To configure input data use the following button: 
The following dialog will appear: 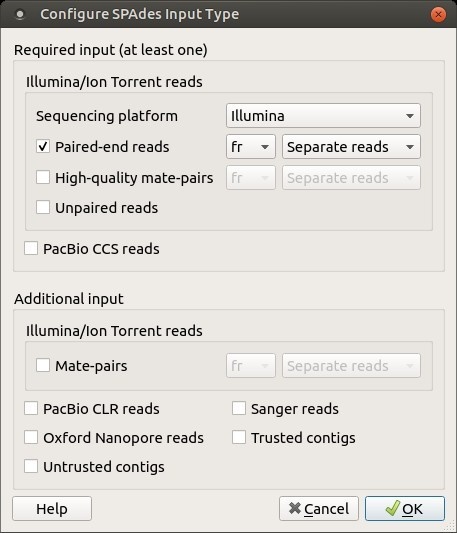
| output-dir | string | |
| Dataset type | Input dataset type. | Multi Cell | dataset-type | string |
| Running mode | Running mode. | Error correction and assembly | running-mode | string |
| K-mers | k-mersizes (-k). | auto | k-mer | numeric |
| Number of threads | Number of threads (-t). | 16 | threads | numeric |
| Memory limit (Gb) | Memory limit (-m). | 250 | memlimit | numeric |
| Output folder | Folder to save Spades output files. | Auto |
Input/Output Ports
The element has 1 input port:
Name in GUI: Spades data
Name in Workflow File: in-data
Slots:
| Slot In GUI | Slot in Workflow File | Type |
|---|---|---|
| URL of a file with right pair reads | url | string |
| URL of a file with reads | url | string |
And 1 output port:
Name in GUI: SPAdes output data
Name in Workflow File: out-data
Slots:
| Slot In GUI | Slot in Workflow File | Type |
|---|---|---|
| ScaffoIds URL | url | string |
| Contig URL | url | string |