To create a new annotation for the active sequence press the Ctrl-N key sequence, select the New annotation toolbar button, or use the Add ‣ New annotation or New annotation context menu item:
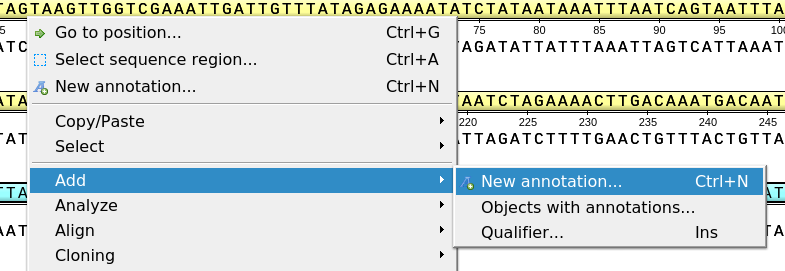
This will activate a dialog where to set up annotation parameters:
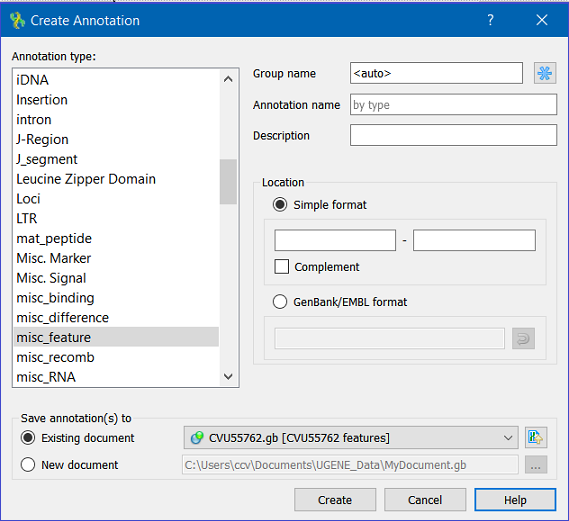
The dialog asks where to save the annotation. It could be either an existing annotation table object or an a new annotation table.
You can also specify the name of the group and the name of the annotation. If the group name is set to <auto> UGENE will use the group name as the name for the group. You can use the ‘/’ characters in this field as a group name separator to create subgroups. If the annotation name is set to by type UGENE will use the annotation type from the Annotation type: table as the name for the annotation. Also, you can add a description in the corresponding text field.
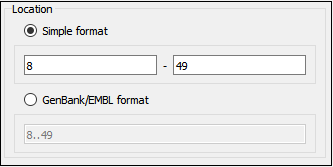
The Location field contains annotation coordinates. The coordinates must be provided in the Genbank or EMBL file formats. If you want to annotate complement strand sequence check the corresponding checkbox for the simple format or surround the coordinates with the “complement()” word or press the last button in the corresponding row to do it automatically.
Note, that by default the Location field contains the coordinates of the selected sequence region.
For amino acid sequences there is no "Complement" check box in the "Simple format" group in the Location section and there is no "Add/remove complement flag" button in the "GenBank/EMBL format" group.
Once the Create button is pressed the annotation is created and highlighted both in the Sequence overview and the Sequence details view areas: