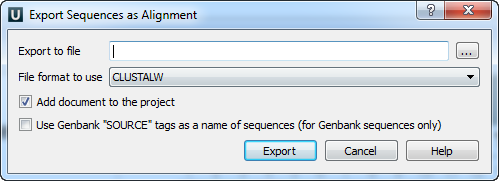
Suppose, we want to interpret FASTA file as multiple alignment. To do this, select a single or several sequence objects in the Project View window, click the right mouse button to open the context menu, and select the Export /Import‣ Export sequences as alignment item:
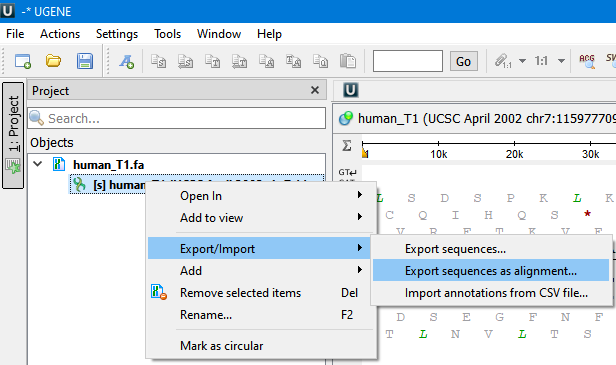
The Export Sequences as Alignment dialog will appear where you can point the result alignment file location, to select a multiple alignment file format, to use Genbank “SOURCE” tags as a name of sequences for Genbank sequences and optionally add the created document to the current project: