In Silico PCR Overview
In silico PCR is used to calculate theoretical polymerase chain reaction (PCR) results using a given set of primers (probes) to amplify DNA sequences.
UGENE provides the In silico PCR feature only for nucleic sequences with Standard DNA and Extended DNA alphabets. To use it in UGENE open a DNA sequence and go to the In silico PCR tab of the Options Panel:
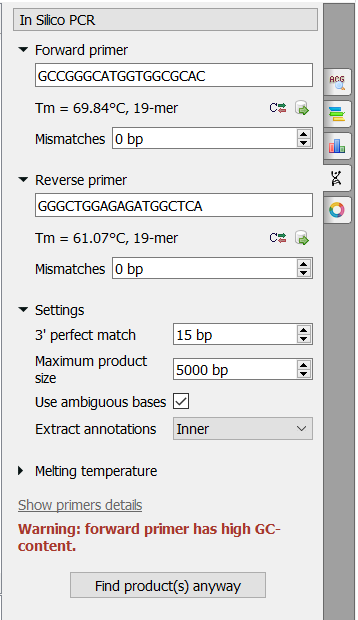
There are the following parameters:
Forward primer - forward primer. The primer length should be between 15 and 50 bases.
Reverse primer - on the opposite strand from the forward primer. The primer length should be between 15 and 50 bases.
Note: you should take in account, that algorithm calculates no more than 50 forward and 50 reverse primers, which means, that you can take 50x50=2500 products maximum.
Mismatches - mismatches limit.
3' perfect match - specify the number of nucleotides at the 3' end that must not have mismatches.
Maximum product size - the maximum size of the amplified sequence. Use ambiguous bases - search for ambiguous bases (as \"N\") if checked.
Extract annotations - specify the type of extracted annotations: Inner, All intersected or None.
- Value Inner is selected by default. When this value is selected, the extracted PCR product contains annotations from the original sequence, located within the extracted region.
- Value All intersected specifies that all annotations of the original sequence that intersect the extracted region must be extracted as well.
- Value None specifies that annotations from the original sequence must not be extracted.
Melting temperature - see the corresponding page for details.
Choosing primers
Type two primers for running In Silico PCR. If the primers pair is invalid for running the PCR process then the warning is shown. Also, primers for the running In silico PCR can be chosen from a primer library. Click the following button to choose a primer from the primers library:
The following dialog will appear:
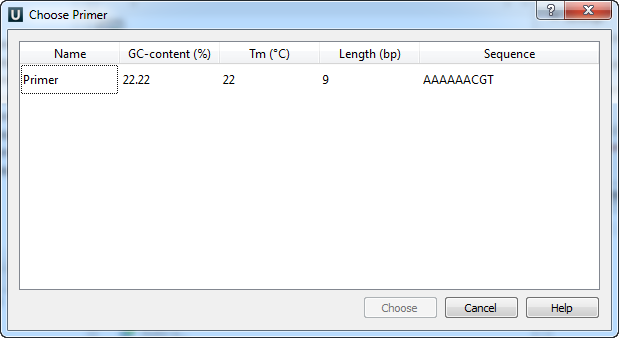
The table consists of the following columns: name, GC-content (%), Tm, Length (bp) and sequence. Select primer in the table and click the Choose button.
Click the Reverse-complement button for making a primer sequence reverse-complement:
Click Show primers details for seeing statistic details about primers.
When you run the process, the predicted PCR products appear in the products table.
Melting temperature
Click the Melting tempeature link for choosing temperature calculation algorithm: Rough or Primer 3.
More information about the temperature options see here: Melting tempeature
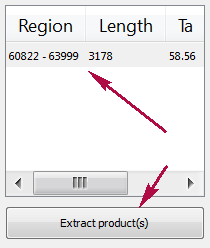
Products table
There are three columns in the table:
- region of product in the sequence
- product length
- preferred annealing temperature
Click the product for navigating to its region in the sequence.
Click the Extract product(s) button for exporting a product(s) in a file or use double click for that.