This workflow simulates the PCR process.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "In Silico PCR" can be found in the "Scenarios" section of the Workflow Designer samples.
Workflow Image
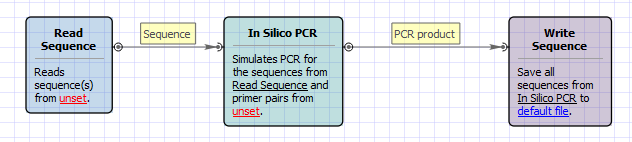
The opened workflow looks as follows:
Workflow Wizard
The wizard has 3 pages.
Input DNA Sequences: On this page you must input DNA sequences.
Primers and Parameters: Here you must input Primers and you can optionally modify In Silico PCR parameters.
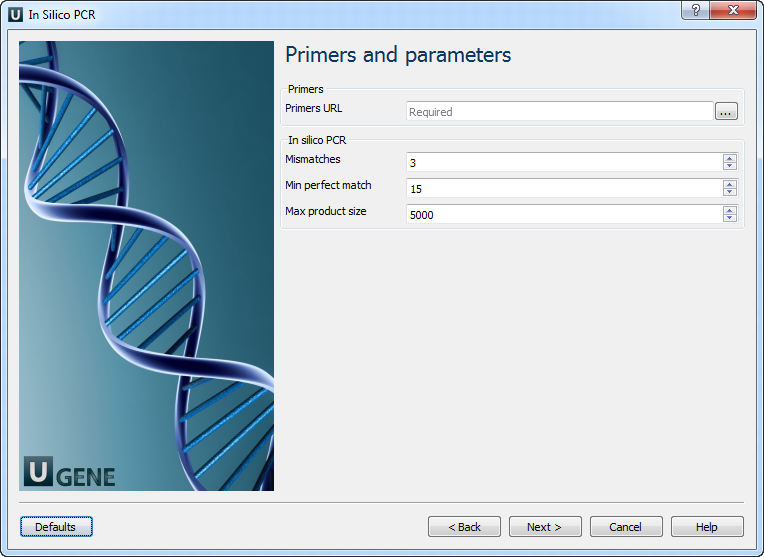
The following parameters are available:
Primers URL A URL to the input file with primer pairs. Mismatches Number of allowed mismatches. Min perfect match Number of bases that match exactly on 3' end of primers. Max product size Maximum size of amplified region. Use ambiguous bases Search for ambiguous bases (as "N") if checked. The input primers file should be in the FASTA format and contain an even number of primers (because each pair should have forward and reverse primers).
Example format:
>forward
CTTGTATGAATGGCCGCACG
>reverse
GATGTAGCGGGTCGTAGTGGOutput data: Here you can see information about output data.