The Phylogenetic Tree Viewer is intended to display a phylogenetic tree built from an alignment or loaded from a file (e.g. a Newick file).
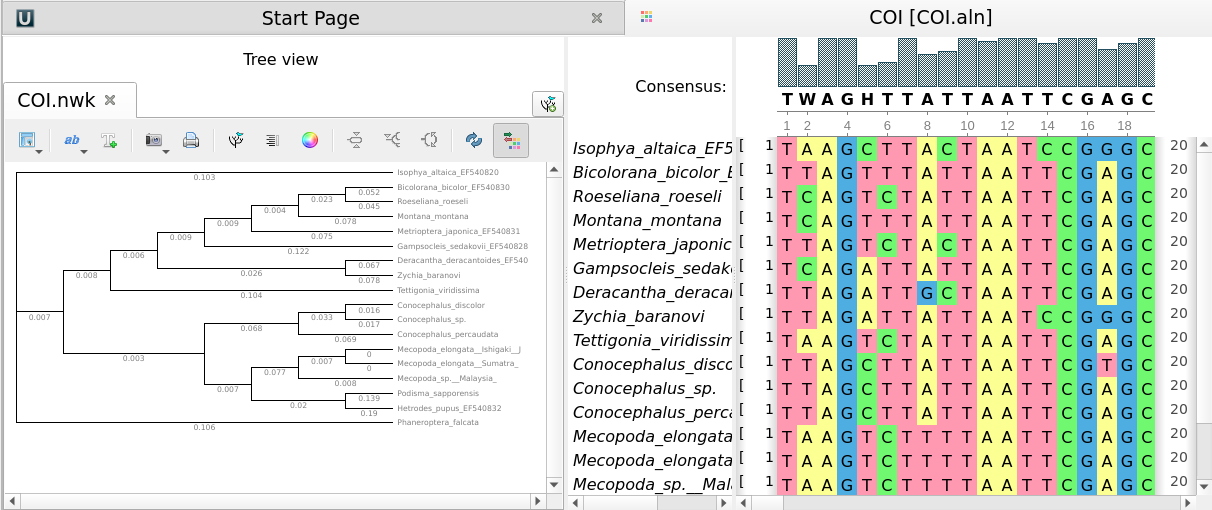
By default, a phylogenetic tree is synchronized with the corresponding alignment.
To disable/enable synchronization use the Tree and Alignment synchronization button.
To load a tree from a file follow the instruction described in the Opening Document paragraph or use the Tree settings tab of the Options Panel. For example, you may open the $UGENE\data\samples\Newick\COI.nwk sample file provided within UGENE package.
To build a tree from a multiple sequence alignment see the Building Phylogenetic Tree paragraph.
To learn what you can do with a tree using UGENE Phylogenetic Tree Viewer read the documentation below.