To apply an alignment highlighting mode, select it in the Highlighting context menu:
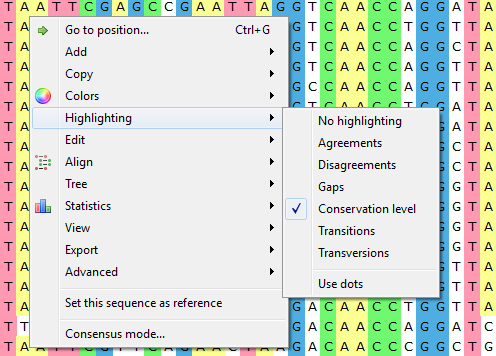
or on the Highlighting tab of the Options Panel:
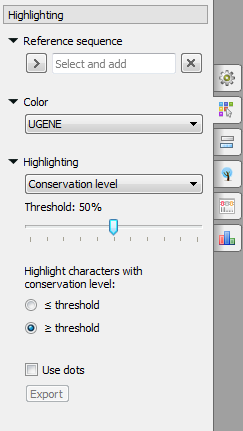
The following modes are available:
Agreements — highlights symbols that coincide with the reference sequence.
Disagreements — highlights nucleotides that differ from the reference sequence.
- Gaps - highlights gaps.
- Conservation level - highlights conservation level of symbols in a multiple alignment >= or <= treshhold. To select the conservation parameters use the Highlighting Options Panel tab.
- Transitions - highlights transitions.
- Transversions - highlights transversions.
To use dots instead of symbols which are not highlighted check the Use dots checkbox in the Options Panel or use the Highlighting->Use dots context menu item.
To select a reference sequence use the Set this sequence as reference context menu or Reference sequence field in the Highlighting tab of the Options Panel.
Also you can export highlighting with a help of the Export button in the Options Panel or by the Export->Export highlighted context menu item. The following dialog will appear:
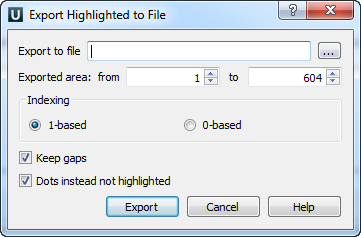
Select file to export, exported area and click on the Export button. The task report will appear in the Notifications.