To create a position weight or frequency matrix from an alignment or a file with several sequences, press the Build new matrix button in the Weight matrix search dialog, or select the Tools ‣ Search for TFBS ‣ Build weight matrix main menu item:
The Build weight or frequency matrix dialog will appear:
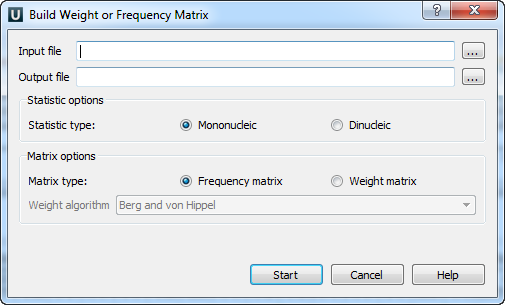
The following parameters are available:
Input file — an alignment or a file with several sequences to build the matrix from. The parameter is mandatory.
Output file — the resulting matrix will be saved in this file. The parameter is mandatory.
Statistic type — defines the way in which the statistics will be collected. The Mononucleic option is basically good for small alignments, and the Dinucleic option must give more appropriate results for big alignments.
Matrix type — defines the type of the resulting matrix.
If the Frequency matrix option is selected then the frequency matrix will be created and saved into the resulting file.
If the Weight matrix option is selected then the intermediate frequency matrix will be created and then transformed into a weight matrix on basis of the selected Weight algorithm. Then the weight matrix will be saved into the resulting file.
For some input files the colored “Alignment Logo” appears at the bottom of the dialog. It gives the representation of the selected alignment.
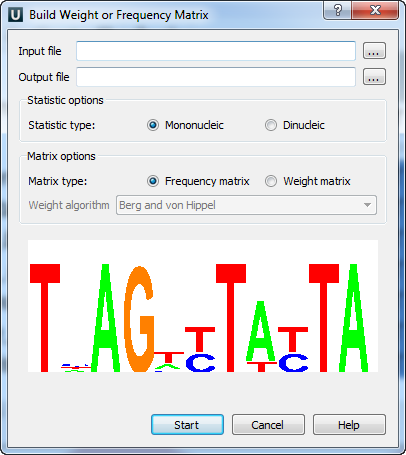
The “Alignment logo” appears when:
- The input file format is *.pfm, *.aln or it is a file with several sequences;
- The size of the input file is small enough.
To start the operation, press the Start button. The matrix will be created and saved. If the Build weight or frequency matrix dialog was invoked from the Weight matrix search dialog, then the matrix also will be chosen as the current profile.