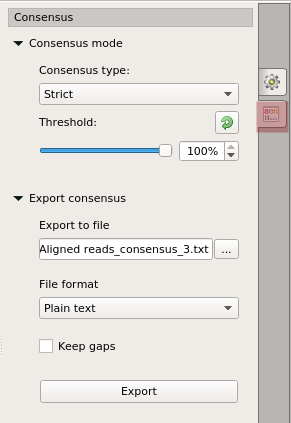
Each base of a consensus sequence is calculated as a function of the corresponding column bases. The Sanger Reads Editor allows switching between different consensus modes: Simple extended and Strict.
The Simple extended algorithm selects the best character from the extended DNA alphabet. Only bases with frequences which are greater than a threshold value are taken into account.
The Strict algorithm returns gap character ('-') if symbol frequency in a column is lower than threshold specified.
To switch the consensus mode go to the Consensus tab of the Options Panel: