The Building Phylogenetic Tree dialog for the PhyML Maximum Likelihood method has the following view:
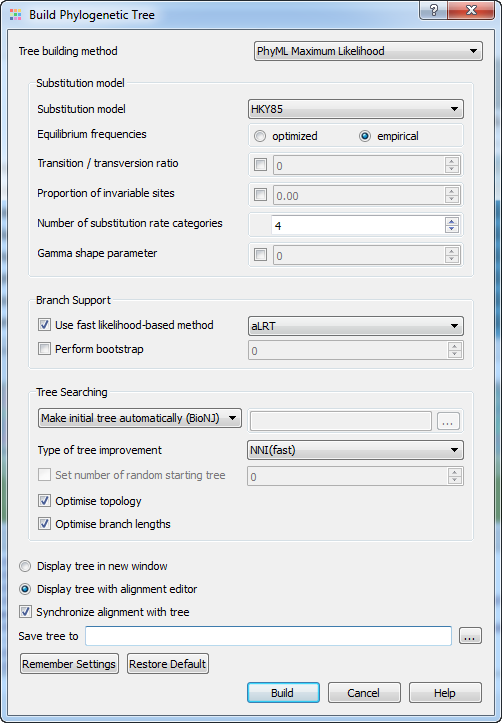
There following parameters are available:
Substitution model parameters:
Substitution model -
Equilibrium frequencies -
Transition/transversion ratio -
Proportion of invariable sites -
Number of substitution rate categories -
Gamma shape parameter -
Branch support parameters:
Use fast likelihood method -
Perform bootstrap -
Tree searching parameters:
Make initial tree automatically -
Type of tree improvement -
Set number of random starting tree -
Optimize topology -
Optimize branch lengths -
Display tree in new window - displays tree in new window.
Display tree with alignment editor - displays tree with alignment editor.
Synchronize alignment with tree - synchronize alignment and tree.
Save tree to - file to save the built tree.
Press the Build button to run the analysis with the parameters selected and build a consensus tree.