This workflow simulates the PCR process.
How to Use This Sample
If you haven't used the workflow samples in UGENE before, look at the "How to Use Sample Workflows" section of the documentation.
Workflow Sample Location
The workflow sample "In Silico PCR" can be found in the "Scenarios" section of the Workflow Designer samples.
Workflow Image
The opened workflow looks as follows:
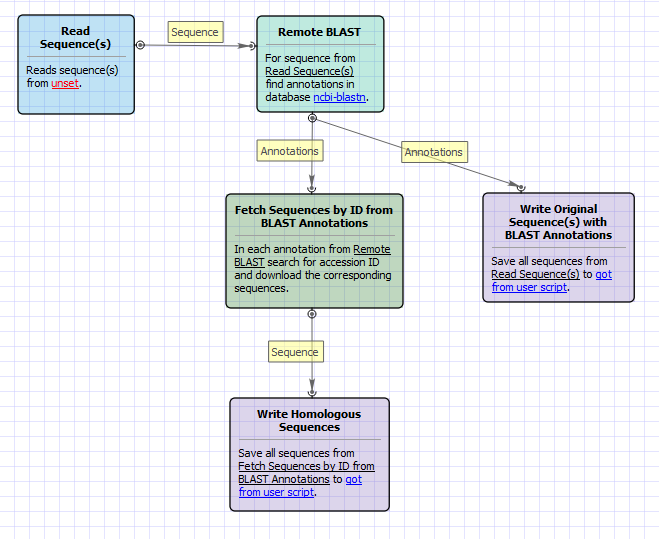
Workflow Wizard
The wizard has 3 pages.
Input Sequence(s) Page: On this page you must input at least one nucleotide sequence.
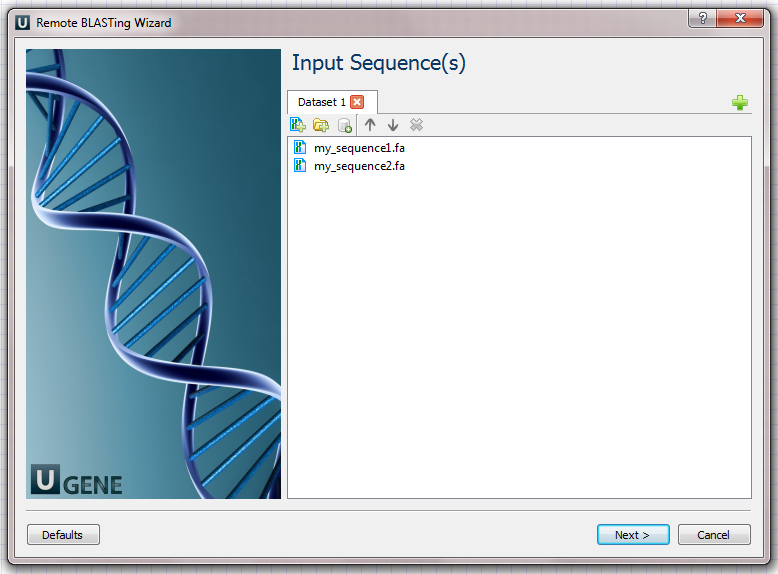
Example Input Data
For example, you can use the following two files as an input to the workflow:
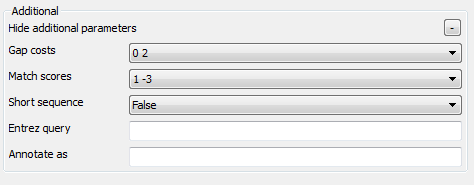
Remote Nucleotide BLAST Page: Here you can optionally modify parameters that should be used for the remote BLAST queries. For example, you can select the search database, correct the e-value and set the maximum number of results (i.e. "Max hits"). The "Megablast" option, applied by default, specifies to optimize the search for high similar sequences only. Selecting it decreases the search time, but some less similar results could be skipped by the search in this case. Note that the "Megablast" option is also applied by default in the NCBI BLAST web interface.
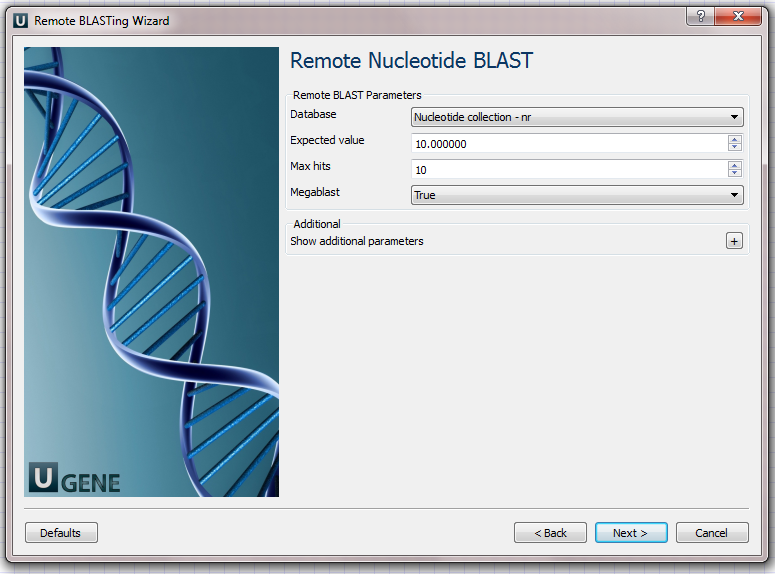
There are also some additional parameters. Description of them can be found in the Remote BLAST workflow element chapter of the documentation.
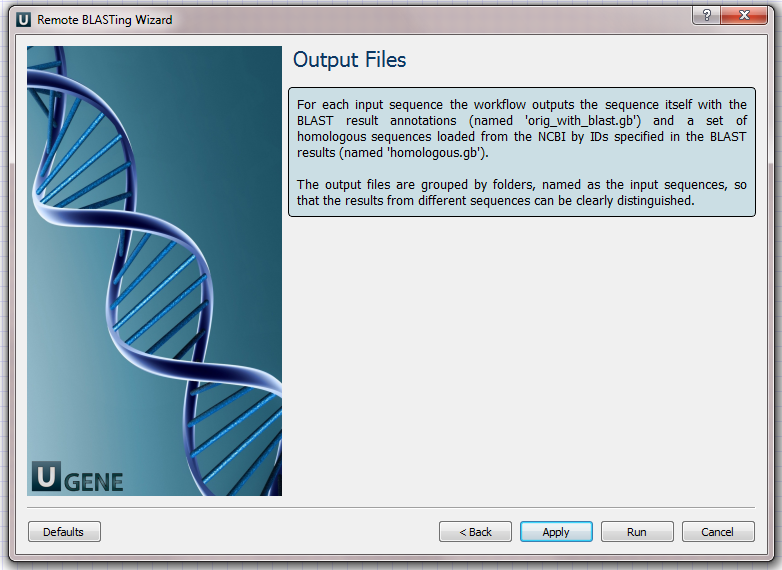
- Output Files Page: this is an informational page. It states that this workflow has predefined names of the output files.
For each input sequence the workflow outputs:- "orig_with_blast.gb" file: the file contains the input sequence itself and the "blast result" annotations;
- "homologous.gb" file: the file contains the found homologous sequences loaded from the NCBI by identifiers, specified in the BLAST results.
The results on the hard drive are grouped by folders (see below).
The wizard page looks as follows: