The Building Phylogenetic Tree dialog for the PhyML Maximum Likelihood method has the following view:
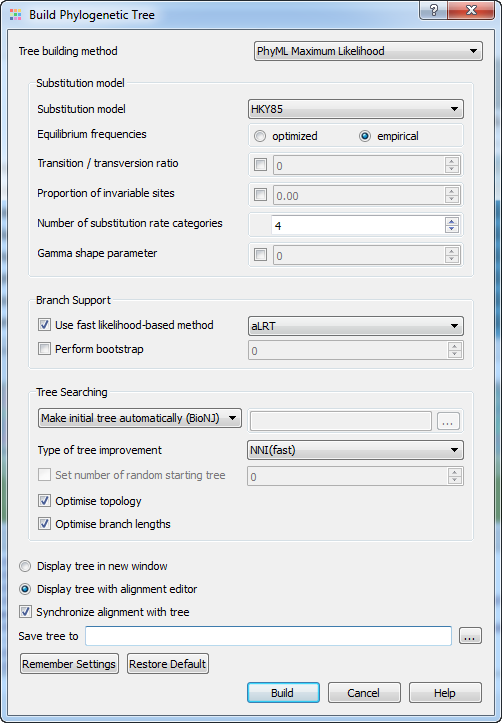
There following parameters are available:
Substitution model parameters:
Substitution model -
Equilibrium frequencies -
Transition/transversion ratio -
Proportion of invariable sites -
Number of substitution rate categories -
Gamma shape parameter -
Branch support parameters:
Use fast likelihood method -
Perform bootstrap -
Tree searching parameters:
Make initial tree automatically -
Type of tree improvement -
Set number of random starting tree -
Optimize topology -
Optimize branch lengths -
Gamma — sets the number of rate categories for the gamma distribution.
You can select the following parameters for the MCMC analisys:
Chain length — sets the number of cycles for the MCMC algorithm. This should be a big number as you want the chain to first reach stationarity, and then remain there for enough time to take lots of samples.
Subsampling frequency — specifies how often the Markov chain is sampled. You can sample the chain every cycle, but this results in very large output files.
Burn-in length — determines the number of samples that will be discarded when convergence diagnostics are calculated.
Heated chains — number of chains will be used in Metropolis coupling. Set 1 to use usual MCMC analysis.
Heated chain temp — the temperature parameter for heating the chains. The higher the temperature, the more likely the heated chains are to move between isolated peaks in the posterior distribution.
Random seed — a seed for the random number generator.
Save tree to — file to save the built tree.
Press the Build button to run the analysis with the parameters selected and build a consensus tree.