Prepare a query schema and save it to a file.
Open a nucleotide sequence that you want to analyze with this query schema. You can see the sequence displayed in the Sequence view.
To learn more about the Sequence view read the main UGENE User Manual.
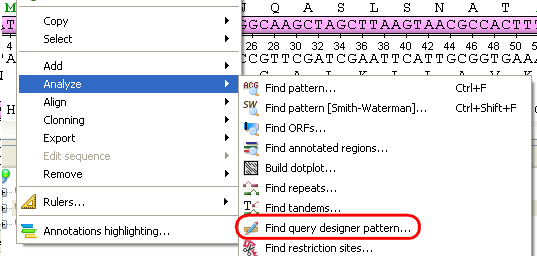
Select the Analyze ‣ Find query designer pattern item in the Actions main menu or in the context menu:
The Analyze with query schema dialog appears:
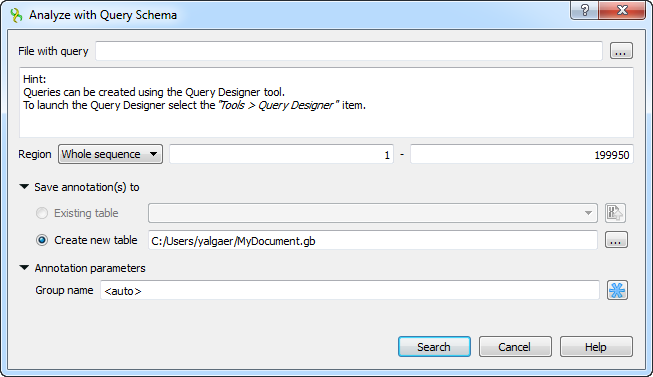
Browse for the file with a query schema. The selected schema preview appears in the dialog, for example:
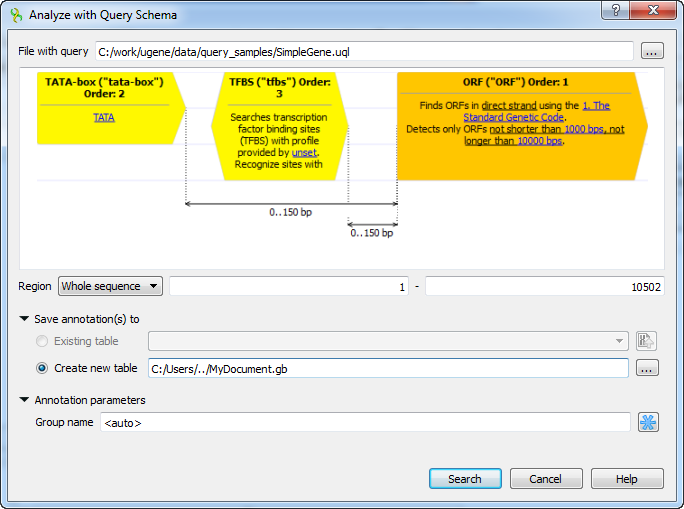
You can also adjust other parameters:
Range — the sequence range to analyze with the query schema, you can select:
- Whole sequence — to analyze the whole sequence.
- Selected range — to analyze the currently selected sequence region. This item is disabled if there is no region selected.
- Custom range — to specify manually a range to analyze.
Save annotation(s) to — specifies where to save the result annotations:
- Existing annotation table — this option is available if there is an existant annotation table and the document is not locked.
- Create new table — saves the result annotations to the specified file in GenBank format.
Group name — name of the annotation group. Note, that the annotation name is set in the query schema file.