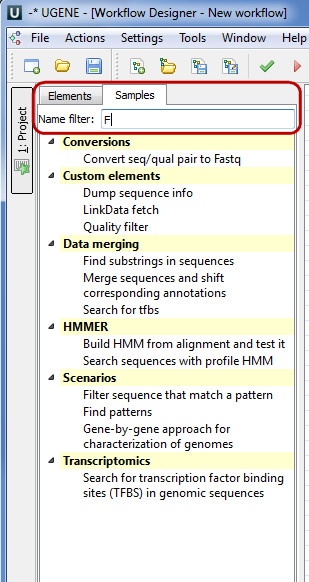
This section contains detailed description of workflow samples presented in the Workflow Designer. To search a sample use the name filter or press the Ctrl+F shortcut that moves you to the name filter also:
- Alignment
- Conversions
- Custom Elements
- Data Marking
- Data Merging
- HMMER
- NGS
- ChIP-Seq Coverage
- ChIP-seq Analysis with Cistrome Tools
- Extract Consensus from Assembly
- Extract Coverage from Assembly
- Extract Transcript Sequences
- Quality Control by FastQC
- De novo Assemble Illumina PE Reads
- De novo Assemble Illumina PE and Nanopore Reads
- De novo Assemble Illumina SE Reads
- De Novo Assembly and Contigs Classification
- Parallel NGS Reads Classification
- Serial NGS Reads Classification
- RNA-Seq Analysis with TopHat and StringTie
- RNA-seq Analysis with Tuxedo Tools
- Variation Annotation with SnpEff
- Call Variants with SAMtools
- Variant Calling and Effect Prediction
- Raw ChIP-Seq Data Processing
- Raw DNA-Seq Data Processing
- Raw RNA-Seq Data Processing
- Get Unmapped Reads
- Sanger Sequencing
- Scenarios
- Transcriptomics
- ChIP-Seq Coverage
- ChIP-seq Analysis with Cistrome Tools
- Extract Consensus from Assembly
- Extract Coverage from Assembly
- Extract Transcript Sequences
- Quality Control by FastQC
- De novo Assemble Illumina PE Reads
- De novo Assemble Illumina PE and Nanopore Reads
- De novo Assemble Illumina SE Reads
- De Novo Assembly and Contigs Classification
- Parallel NGS Reads Classification
- Serial NGS Reads Classification
- RNA-Seq Analysis with TopHat and StringTie
- RNA-seq Analysis with Tuxedo Tools
- Variation Annotation with SnpEff
- Call Variants with SAMtools
- Variant Calling and Effect Prediction
- Raw ChIP-Seq Data Processing
- Raw DNA-Seq Data Processing
- Raw RNA-Seq Data Processing
- Get Unmapped Reads